cytodatagen
A Python package for generating synthetic flow cytometry data
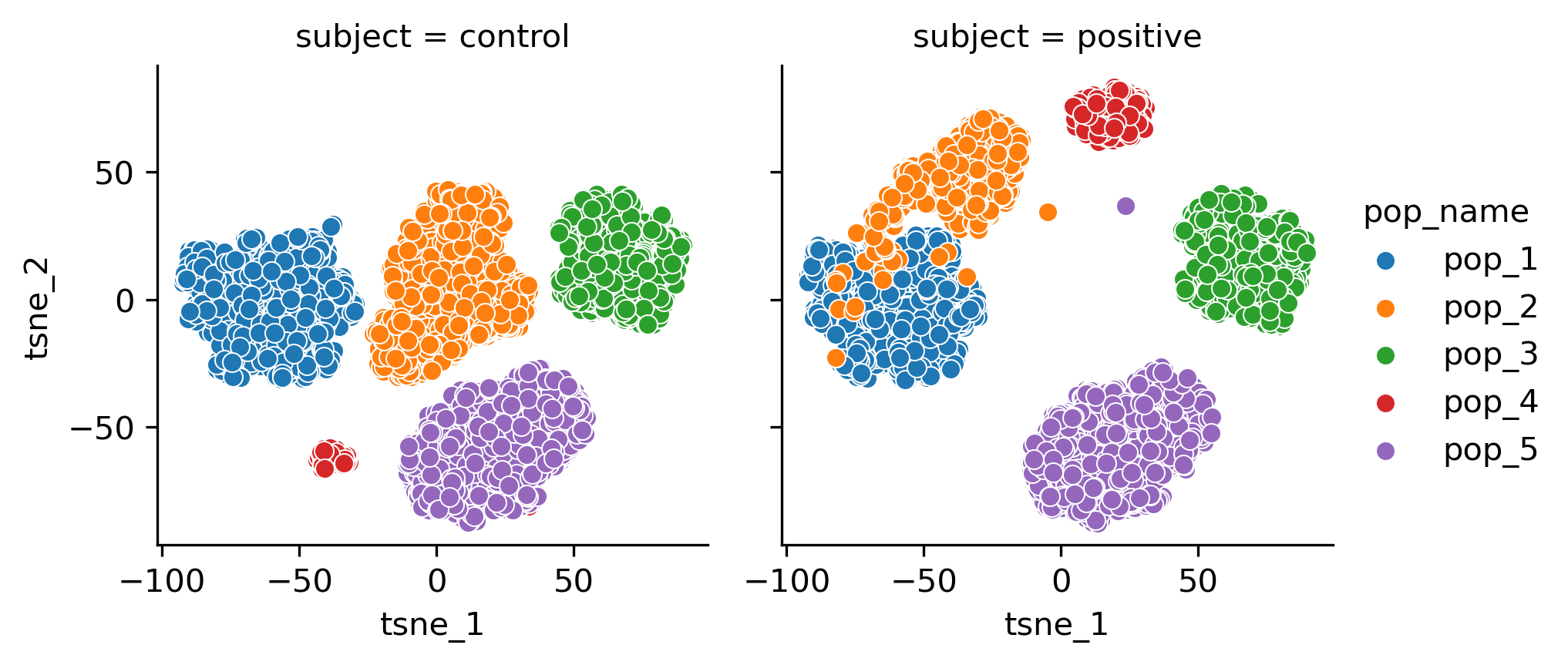
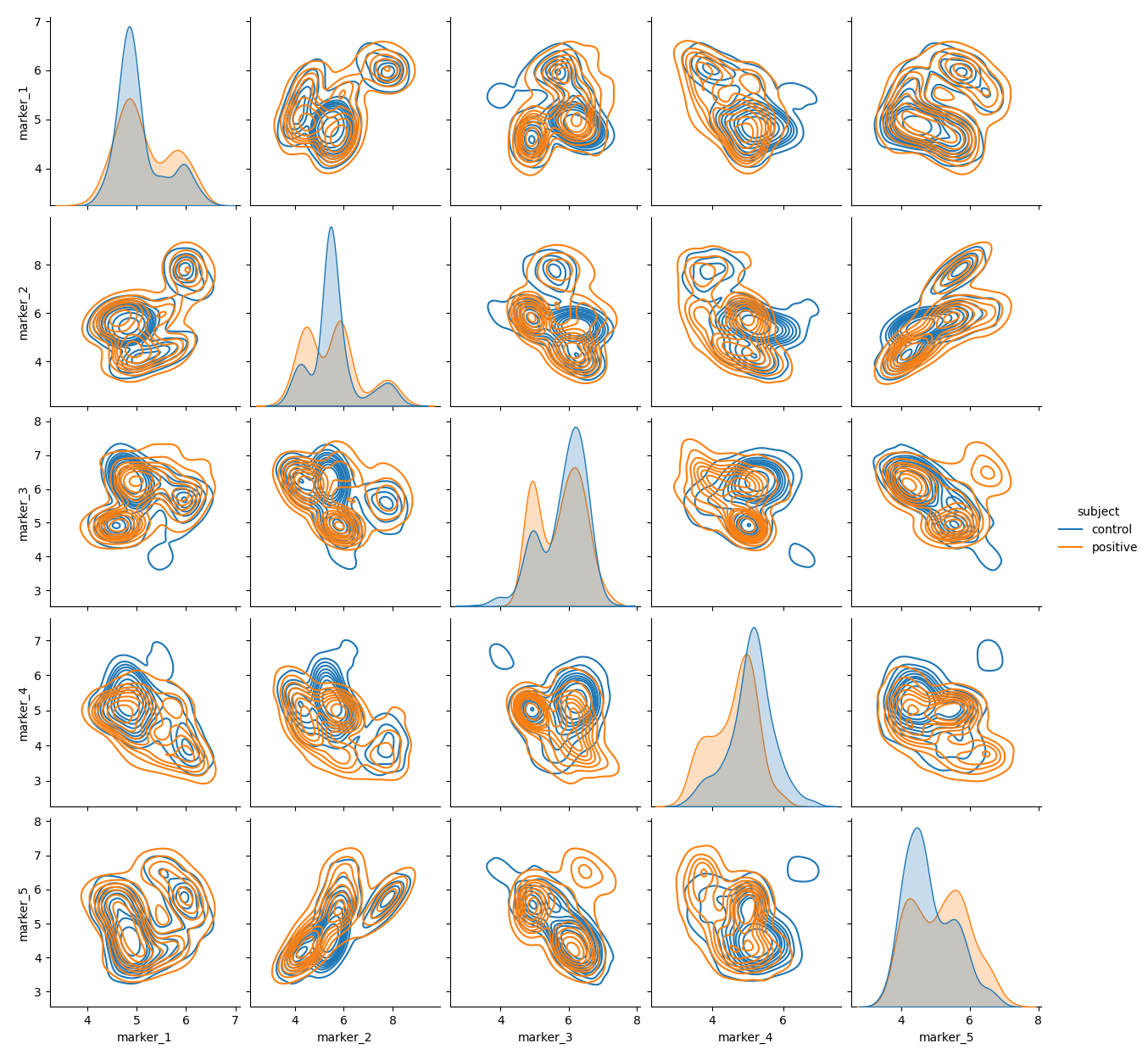
Synthetic datasets are useful to validate machine learning algorithms across varying dataset properties. We present cytodatagen, a Python package for generating synthetic flow cytometry and CyTOF data. Fluoroessence signals are modelled as a mixture of multivariate normal distributions. However, a modular design allows for rapid extensions, like other probability distributions. cytodatagen supports data export to the Fow Cytometry Standard format, as well as the scverse-central AnnData format. The code is available at: https://github.com/bckrlab/cytodatagen