Evaluating the Fidelity of Explanations for Convolutional Neural Networks in Alzheimer's Disease Detection
XAI for Alzheimer's Disease detection
Can we trust explanations of neural network decisions in Alzheimer’s Disease (AD) detection?
Key Findings
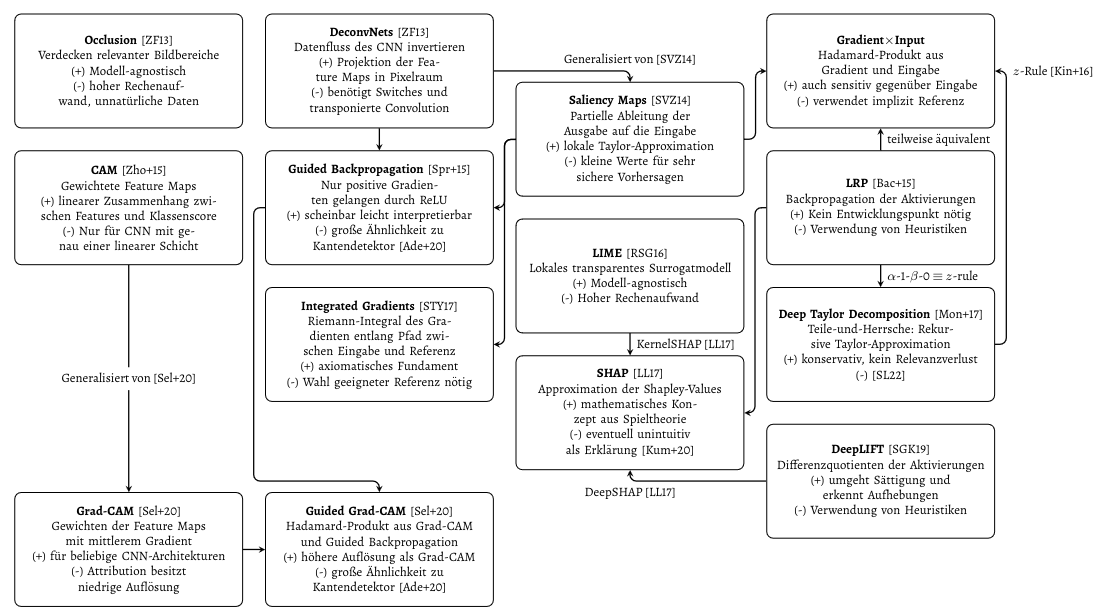
A plethora of explanation methods for convolutional neural networks is available, each with individual benefits and drawbacks.
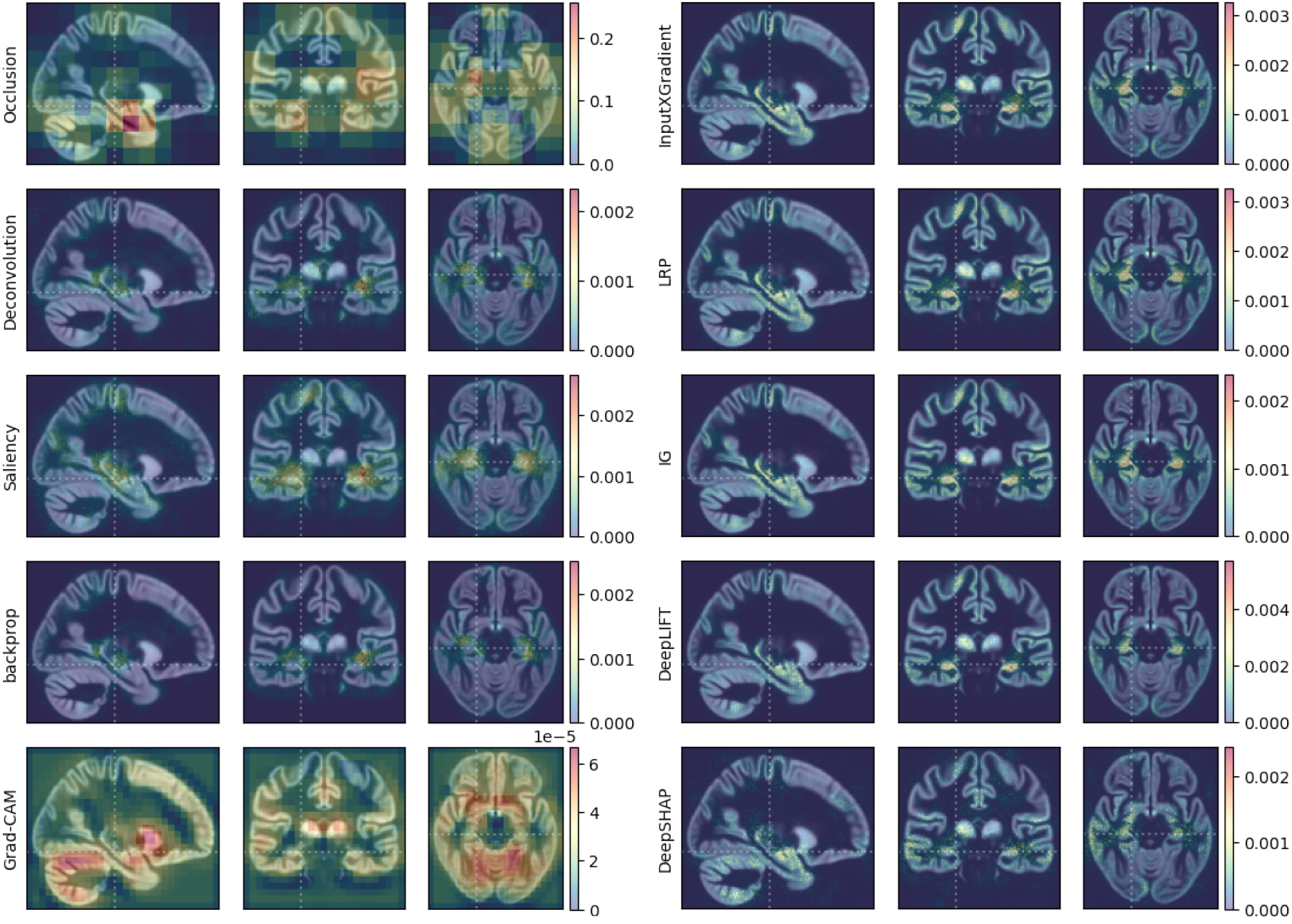
Even though generated attribution maps may appear similar by visual inspection, they can differ significantly regarding the fidelity towards the underlying classifier.**
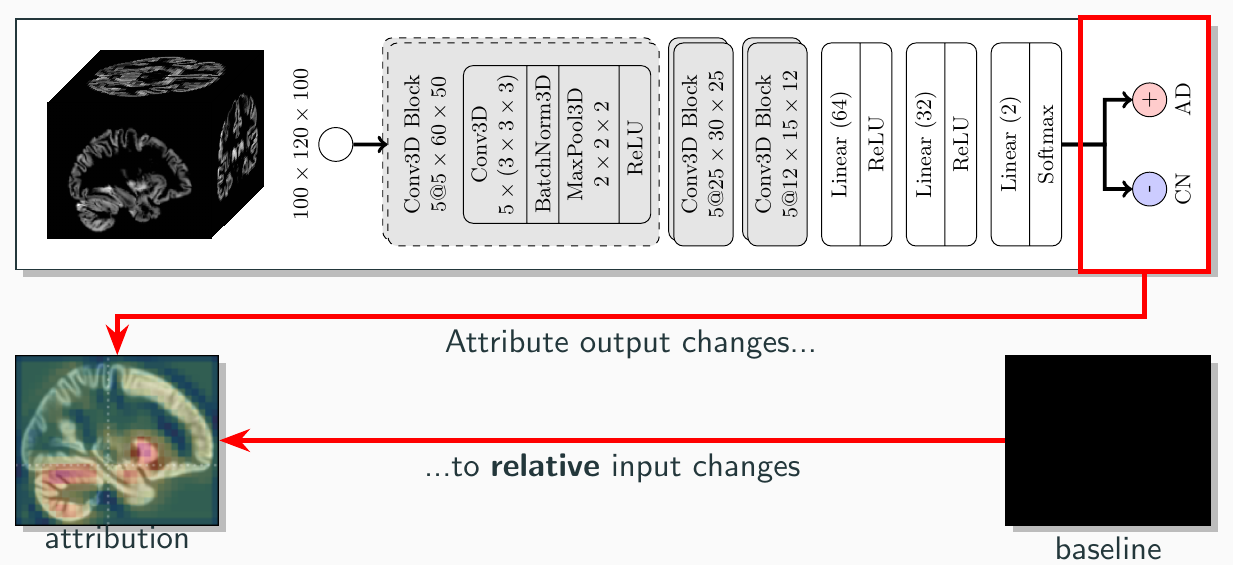
 To evaluate the fidelity of the explanation methods, we employed the perturbation based *deletion* metric.
To evaluate the fidelity of the explanation methods, we employed the perturbation based *deletion* metric. 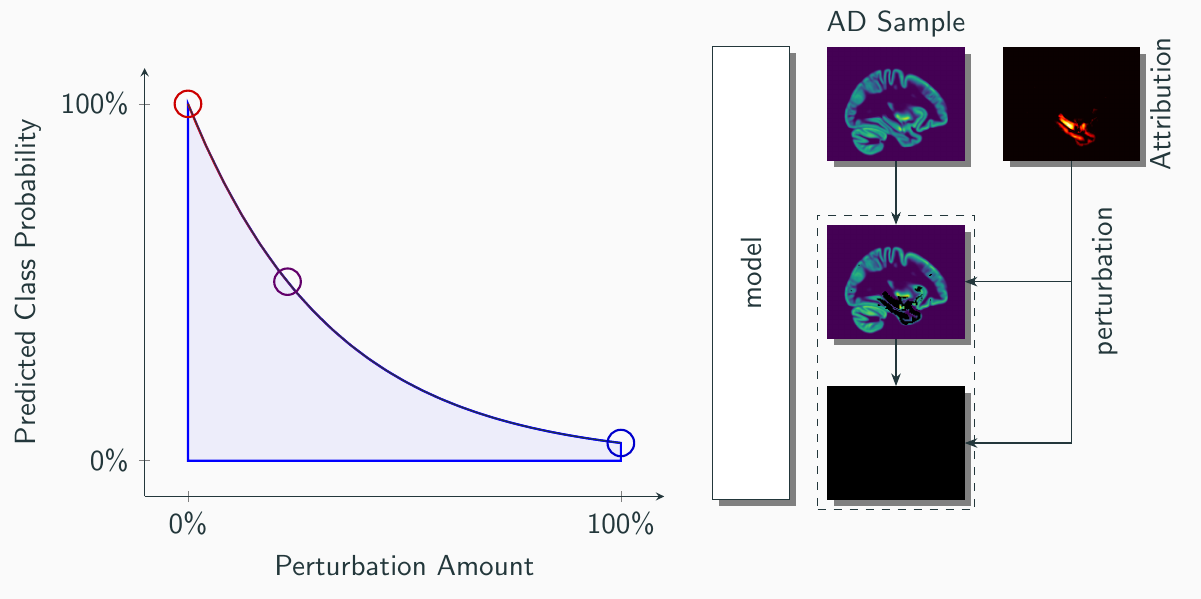 Interestingly, this revealed differences between the explanation methods in their ability to decrease the predicted probability for AD:
Interestingly, this revealed differences between the explanation methods in their ability to decrease the predicted probability for AD: 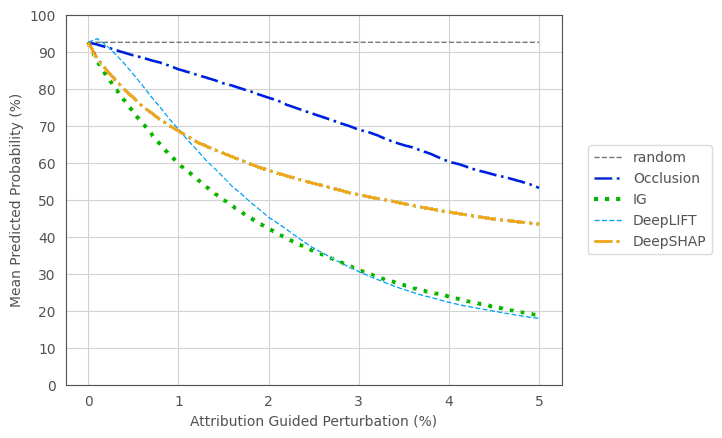
Interpretation of attribution maps should always include the employed reference image, since its selection allows generating an almost arbitrary explanation.
 For example, we observed explanation methods to assign the hippocampal area negative contribution for classifying a AD sample with AD - Even though hippocampal atrophy is a gold standard biomarker in AD detection. We hypothesize, that the neural network learned to utilize atrophy patterns, i.e., areas with reduced signal strength, to classify AD. However, since a null image was used as reference to generate explanations, even the atrophied AD hippocampus had more overall signal then the null image. Thus, the atrophied AD hippocampus was *decreasing* the probability for Alzheimer's Disease *relative to the reference image*. Therefore, the explanation method ended up assigning negative contribution.
For example, we observed explanation methods to assign the hippocampal area negative contribution for classifying a AD sample with AD - Even though hippocampal atrophy is a gold standard biomarker in AD detection. We hypothesize, that the neural network learned to utilize atrophy patterns, i.e., areas with reduced signal strength, to classify AD. However, since a null image was used as reference to generate explanations, even the atrophied AD hippocampus had more overall signal then the null image. Thus, the atrophied AD hippocampus was *decreasing* the probability for Alzheimer's Disease *relative to the reference image*. Therefore, the explanation method ended up assigning negative contribution. In conclusion, we found the mean image of the control class to generate more interpretable explanations, since it asks the question:
“Why was this subject predicted to have AD - compared to a healthy control subject?”
Conference Talk
The results have been presented at the German Conference on Medical Image Computing (BVM) 2025:
Acknowledgements
Funding was provided by the BMBF (01IS22077) and DFG (DY151/2-1). Data was provided by the Alzheimer’s Disease Neuroimaging Initiative (ADNI).
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.

