DeepCell
Bachelor & Master Thesis
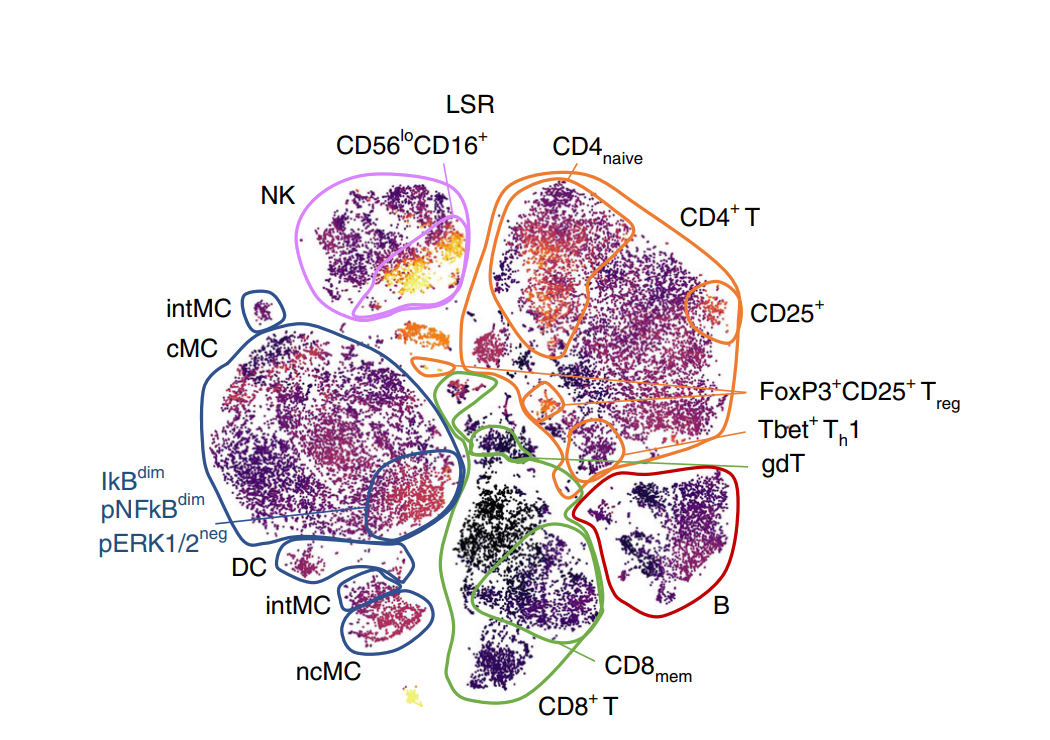
Modern technologies allow characterization of millions of individual cells per patient, and provide for an accurate understanding of complex biological systems by providing insights into cellular heterogeneity and novel cellular subsets. However, these high-dimensional data are traditionally analyzed by gating on bivariate dot plots, which are not only laborious given the quadratic increase of complexity with dimension but are also biased through this manual process. This can adversely affect downstream analyses and predictions. To address this, deep learning methods have shown potential to directly work on single cell data to produce highly accurate predictions in application scenarios like diagnosing the latent cytomegalovirus (CMV) in healthy individuals. Nevertheless, these approaches are only recently emerging and it is necessary to assess their performance on the multitude of available problem settings.
In this project, the goal is to evaluate the performance of deep learning methods for single-cell data in multiple predictive settings. This includes existing algorithms as well as novel approaches based on geometric deep learning. Tasks for evaluation include pregnancy related settings like preterm and preeclampsia prediction, healing processes, and more. Possible extensions of the project are novel, task-specific visualization algorithms as well as the integration of background knowledge for better predictive performance and deeper insights into the underlying biology.