Trees and Forests#
Building a simple Decision Tree Classifier!
Author: Bjarne C. Hiller
Show code cell content
import numpy as np
import pandas as pd
from sklearn.base import BaseEstimator, ClassifierMixin
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
Ingredients#
np.uniqueandnp.unique_countsnp.argmax
Implementation#
Let’s start with a simple class to hold the decision rules associated with each tree node. Each decision rule splits the data by comparing one of the features against a threshold value.
class TreeNode:
"""Represents one split in the Decision Tree."""
def __init__(self, feature=None, value=None):
self.left = None
self.right = None
self.feature = feature
self.value = value
self.samples = None
def split(self, X):
left = X[..., self.feature] < self.value
return left, ~left
def is_leaf(self):
"""Is this a leaf node?"""
return (self.left is None) and (self.right is None)
def __repr__(self):
if self.is_leaf():
return f"TreeNode({self.samples})"
return f"TreeNode({self.samples}, {self.feature} < {self.value:.2f})"
Next, we will write a class that generates split candidates. One of the simplest approaches is to use the median of each feature as split value:
class Splitter:
def make_splits(self, X, y) -> list[TreeNode]:
"""Generates candidate splits."""
pass
class MedianSplitter(Splitter):
"""Generates splits by using the Median for each feature."""
def make_splits(self, X, y) -> list[TreeNode]:
n, p = X.shape
splits = []
medians = np.median(X, axis=0)
for i in range(p):
node = TreeNode(feature=i, value=medians[i])
splits.append(node)
return splits
Now, we need some criterion to evaluate the generated splits and pick the best candidate:
class Criterion:
def evaluate(self, X, y, split: TreeNode):
pass
def __call__(self, X, y, split: TreeNode):
return self.evaluate(X, y, split)
The Gini index, named after the mathematician Corrado Gini, is a measure of class impurity, that the Decision Tree tries to minimize. It represents, of how often a randomly chosen instance would be incorrectly labeled, if labels were decided randomly according to the relative number of instances of the respective class.
Let
Then, the gini index
A gini index of 0 means, that all instances belong to the same class.
def gini_index(labels) -> float:
"""Computes the Gini-Index for a given numpy array of labels."""
gini = 0
# TODO: Implement computation of the Gini-Index!
return gini
Show code cell content
def gini_index(labels) -> float:
"""Computes the Gini-Index for a given numpy array of labels."""
_, counts = np.unique_counts(labels)
p = counts / counts.sum()
return 1 - np.power(p, 2).sum()
assert gini_index([0,0,0]) == 0
assert gini_index([0,1]) == 0.5
class GiniCriterion(Criterion):
"""Evaluates splits using the gini index."""
def evaluate(self, X, y, split: TreeNode):
left, right = split.split(X)
gini = gini_index(y)
gini_left = 0
if len(y[left]) > 0:
gini_left = len(y[left]) / len(y) * gini_index(y[left])
gini_right = 0
if len(y[right]) > 0:
gini_right = len(y[right]) / len(y) * gini_index(y[right])
return gini - gini_left - gini_right
Finally, we will bring everything together in this Tree class. At each node, the tree will generate new candidate splits with the Splitter object, evaluate them with the criterion, and use the best candidate to split the data into a left and a right child node. This will continue, until all samples belong to the same class or the maximum depth is reached:
class Tree(BaseEstimator, ClassifierMixin):
def __init__(self, max_depth=None, criterion="gini", splitter="median"):
super().__init__()
self.max_depth = max_depth
self.root_ = None
self.classes_ = None
self.criterion = criterion
if self.criterion == "gini":
self.criterion = GiniCriterion()
self.splitter = splitter
if self.splitter == "median":
self.splitter = MedianSplitter()
def fit(self, X, y):
# save class labels observed in training data
self.classes_ = np.unique(y)
# recursively grow tree and return root node
self.root_ = self.grow(X, y, 1)
def class_counts(self, y):
"""Helper function to receive class counts."""
bins = np.zeros_like(self.classes_)
for i, c in enumerate(self.classes_):
bins[i] = np.sum(y == c)
return bins
def grow(self, X, y, depth):
"""Recursively grow a tree."""
n, p = X.shape
samples = self.class_counts(y)
# return leaf node
if len(np.unique(y)) <= 1 or (self.max_depth is not None and depth >= self.max_depth):
leaf = TreeNode()
leaf.samples = samples
return leaf
# generate splits
splits = self.splitter.make_splits(X, y)
# evaluate splits and aggregate scores
scores = [self.criterion.evaluate(X, y, split) for split in splits]
# gest best split
node = splits[np.argmax(scores)]
node.samples = samples
left, right = node.split(X)
# check for degenerate splits
if len(y[left]) == 0 or len(y[left]) == 0:
leaf = TreeNode()
leaf.samples = samples
return leaf
# grow left and right children
node.left = self.grow(X[left], y[left], depth+1)
node.right = self.grow(X[right], y[right], depth+1)
return node
def traverse(self, x) -> TreeNode:
node = self.root_
while not node.is_leaf():
left, right = node.split(x)
if left.all():
node = node.left
else:
node = node.right
return node
def predict(self, X):
predictions = []
for x in X:
# get leaf nod
leaf = self.traverse(x)
main_class = np.argmax(leaf.samples)
y = self.classes_[main_class]
predictions.append(y)
return predictions
def __str__(self):
text = ""
nodes = [self.root_]
while nodes:
text += "\t".join([str(n) for n in nodes]) + "\n"
nodes = sum([[n.left, n.right] for n in nodes], start=[])
nodes = [n for n in nodes if n is not None]
return text
Test on Iris Dataset#
Let’s try out our implementation on the iris dataset:
iris_ds = load_iris()
# only use petal length and petal width
X = iris_ds["data"][:, [2,3]]
y = iris_ds["target"]
X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.8, random_state=19)
tree = Tree()
tree.fit(X_train, y_train)
tree.root_
TreeNode([40 38 42], 0 < 4.45)
from sklearn.metrics import accuracy_score
y_pred = tree.predict(X_test)
acc = accuracy_score(y_test, y_pred)
print(f"Test Accuracy: {acc:.2f} %")
Test Accuracy: 0.97 %
tree
TreeNode([40 38 42], 0 < 4.45) TreeNode([40 20 0], 0 < 1.55) TreeNode([ 0 18 42], 1 < 1.80) TreeNode([30 0 0]) TreeNode([10 20 0], 1 < 1.00) TreeNode([ 0 17 4], 1 < 1.50) TreeNode([ 0 1 38], 1 < 2.00) TreeNode([10 0 0]) TreeNode([ 0 20 0]) TreeNode([0 5 0]) TreeNode([ 0 12 4], 0 < 4.65) TreeNode([ 0 1 14], 0 < 5.10) TreeNode([ 0 0 24]) TreeNode([0 7 1]) TreeNode([0 5 3], 0 < 5.00) TreeNode([0 1 5], 0 < 4.85) TreeNode([0 0 9]) TreeNode([0 3 0]) TreeNode([0 2 3], 1 < 1.60) TreeNode([0 1 2]) TreeNode([0 0 3]) TreeNode([0 0 2]) TreeNode([0 2 1], 0 < 5.10) TreeNode([0 1 0]) TreeNode([0 1 1], 0 < 5.45) TreeNode([0 1 0]) TreeNode([0 0 1])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
TreeNode([40 38 42], 0 < 4.45) TreeNode([40 20 0], 0 < 1.55) TreeNode([ 0 18 42], 1 < 1.80) TreeNode([30 0 0]) TreeNode([10 20 0], 1 < 1.00) TreeNode([ 0 17 4], 1 < 1.50) TreeNode([ 0 1 38], 1 < 2.00) TreeNode([10 0 0]) TreeNode([ 0 20 0]) TreeNode([0 5 0]) TreeNode([ 0 12 4], 0 < 4.65) TreeNode([ 0 1 14], 0 < 5.10) TreeNode([ 0 0 24]) TreeNode([0 7 1]) TreeNode([0 5 3], 0 < 5.00) TreeNode([0 1 5], 0 < 4.85) TreeNode([0 0 9]) TreeNode([0 3 0]) TreeNode([0 2 3], 1 < 1.60) TreeNode([0 1 2]) TreeNode([0 0 3]) TreeNode([0 0 2]) TreeNode([0 2 1], 0 < 5.10) TreeNode([0 1 0]) TreeNode([0 1 1], 0 < 5.45) TreeNode([0 1 0]) TreeNode([0 0 1])
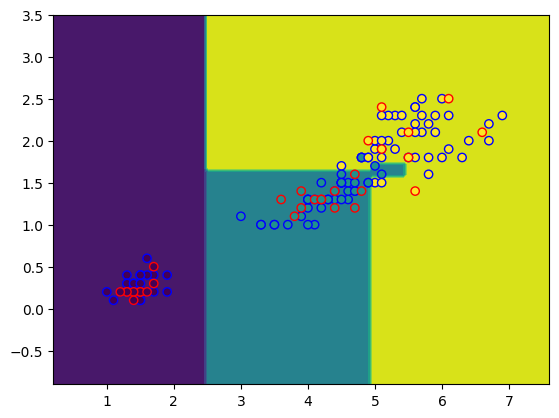
from sklearn.inspection import DecisionBoundaryDisplay
import matplotlib.pyplot as plt
DecisionBoundaryDisplay.from_estimator(tree, X_test)
plt.scatter(X_train[:,0], X_train[:,1], c=y_train, edgecolors="b")
plt.scatter(X_test[:,0], X_test[:,1], c=y_test, edgecolors="r");
Outlook#
Fortunately, sklearn already provides an implementation via the DecisionTreeClassifier and DecisionTreeRegressor
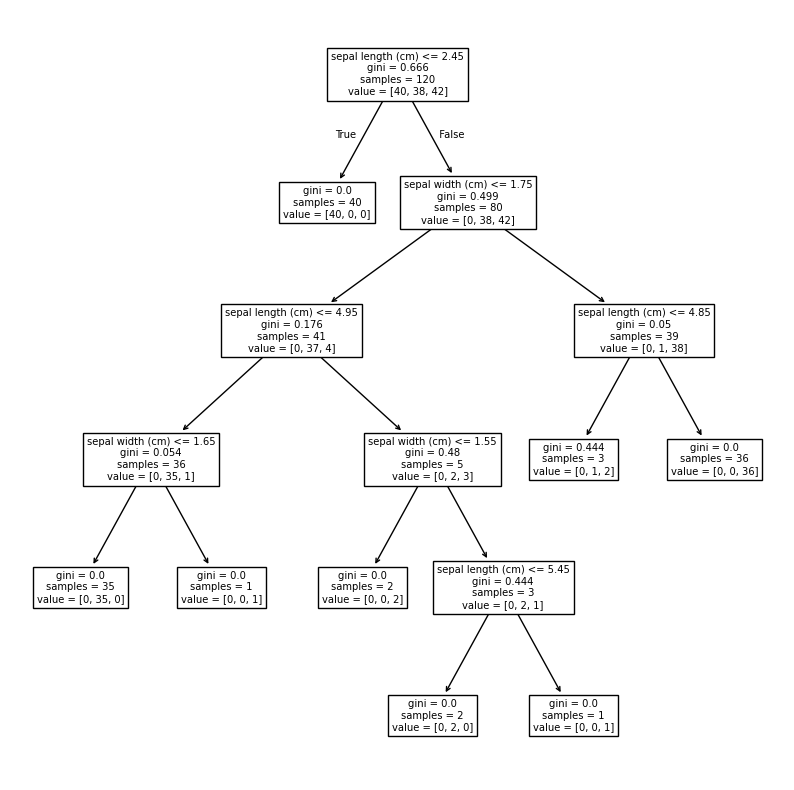
from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor, plot_tree
model = DecisionTreeClassifier()
fig, ax = plt.subplots(figsize=(10,10))
model.fit(X_train, y_train)
plot_tree(model, feature_names=iris_ds["feature_names"], ax=ax);
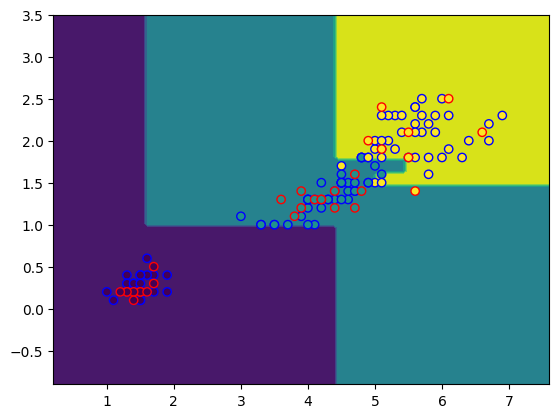
fig, ax = plt.subplots()
DecisionBoundaryDisplay.from_estimator(model, X_test, ax=ax)
plt.scatter(X_train[:,0], X_train[:,1], c=y_train, edgecolors="b")
plt.scatter(X_test[:,0], X_test[:,1], c=y_test, edgecolors="r");